Current Research Projects in the Li Group
Investigating the Multiscale Beauty of Chemistry
Our research focuses on the innovation and applications of multiscale modeling to understand complex chemical systems. The molecules that we study can be as small as a molecular wrench, or as big as a superhelical fiber. By examining a wide range of length and time scales, we strive to elucidate the critical structure-mechanism-function relationships of these molecules, and provide rational guide to help drug discovery and materials design.
Quick links to the brief introduction of each research direction and related publications.
Multiscale Theory for Complex Systems: Computational Microscope to Zoom In and Out
Despite the advance of modern computers and computational technology, the current capability to study complex systems is limited to microscopic length and time scales. To explore the multiscale complexity, we are developing new theory and methodology to model large biological/material systems and long chemical processes. One of our basic ideas is to adapt the model resolutions — to zoom in and out — based on the needs of sampling, while smart mapping and reverse mapping approaches help smooth transitions between models of different resolutions.

Peptide Aggregation and Activation: Magic of Simplicity and Diversity
Despite the simplicity in sequence and structure, peptides display amazing properties and functions. We are interested in oligopeptides and their self-assembled nanostructures; we are fascinated by peptides that can aggregate and form pores in membranes for treating antibiotic resistance and cancers; we also study neuropeptides and their crucial physiological roles. Our mutliscale modeling technique allows to to explore the large chemical space accurately and efficiently, and develop predictive tools for the design of antimicrobial/anticancer agents and biocompatible nanomaterials.

Targeting New GPCRs and Kinases: From Computer Design to Bedside
G protein-coupled receptors (GPCRs) and kinases probably represent the most important protein targets for drug discovery. Many drugs act on them directly or their signaling pathways indirectly. To guide rational drug design, reliable computational methods and models need to be developed. We are employing multiscale modeling and simulation approaches to investigate the structure-mechanism relationship of these proteins. Particularly, we focus on the GPCRs and kinases that have multiple domains and undetermined full-length structures.

Rational Design of DNA Nanodevices: Use Codes of Life to Program Biocompatible Materials
DNA nanostructures that are formed by minimal numbers of DNA strands hold practical promise in material and medical applications. We have developed a software program to systematically construct DNA nanostructures like cages and junctions, with unlimited chemical modifications. These nanostructures display surprising dynamics and functions, which are being studied with our innovative multiscale modeling strategies.
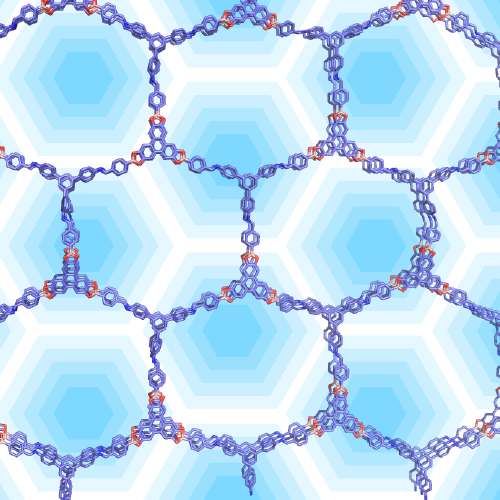
Rational Design of Organic Molecular Devices: Well-Defined Shapes, Structures, and Advanced Functions
Discovery of organic molecular devices is no longer serendipity. With the help from multiscale modeling, we can model the design and predict potential properties of new materials. Currently, we are studying new organic molecules and structures with well defined shapes. It is our goal to create systematic multiscale modeling approaches to guide the design and synthesis of these materials.