About me

I have been conducting studies on high-throughput cytometry, high-content biological imaging and analysis, biological pattern recognition, and applications of statistical machine learning in cell biology, cancer research, neuroscience, bioengineering, and agricultural engineering. My research’s underlying theme is the use of innovative data-science tools to reduce complex phenotypic information into essential statistical models and gain insight into biological processes characterized by incomplete and noisy data.
One project that exemplifies the highly interdisciplinary nature of this inquiry and the inherently collaborative nature of my research is the development of a non-exhaustive learning framework (a set of methods addressing problems of learning with complex, noisy, and highly incomplete training data).
The work, which was initially inspired by a practical pathogen-detection problem (research project sponsored by USDA-ARS) led to the development of a unique feature-extraction system capable of quantifying phenotypic changes in bacterial colonies. Subsequently, this research agenda expanded to address a broader issue of detection and/or classification of bacterial phenotypes in the absence of a complete model of the data source. The methodology created in collaboration with Dr. M Murat Dundar (IUPUI) initially relied on a density-based approach, which used class-conditional likelihoods to detect new entities in samples. The success of this design encouraged further investigation, and the study was expanded to deliver a first universal paradigm for learning with incomplete or partially observed information via non-parametric Bayesian self-adjusting models. Clinical practitioners have recognized the potential of this strategy, and the technique has been successfully applied for the automated diagnosis of acute myeloid leukemia (AML) and the prediction of disease progression.
The described study seamlessly integrated the components of machine learning, statistics, microbiology, cell biology, and immunology. It demonstrated the crucial role of data science in modern biomedical research and illustrated the importance of multidisciplinary approaches reaching beyond established disciplines and narrowly defined fields. I firmly believe that his ability to translate complex problems of quantifying biological responses into reducible mathematical constructs not only allows me to pursue an original and highly innovative research agenda but also enables me to serve as a key scientific intermediary and idea conduit between diverse groups of collaborators approaching the problems from the computational and biological perspectives.
My publication list is here.
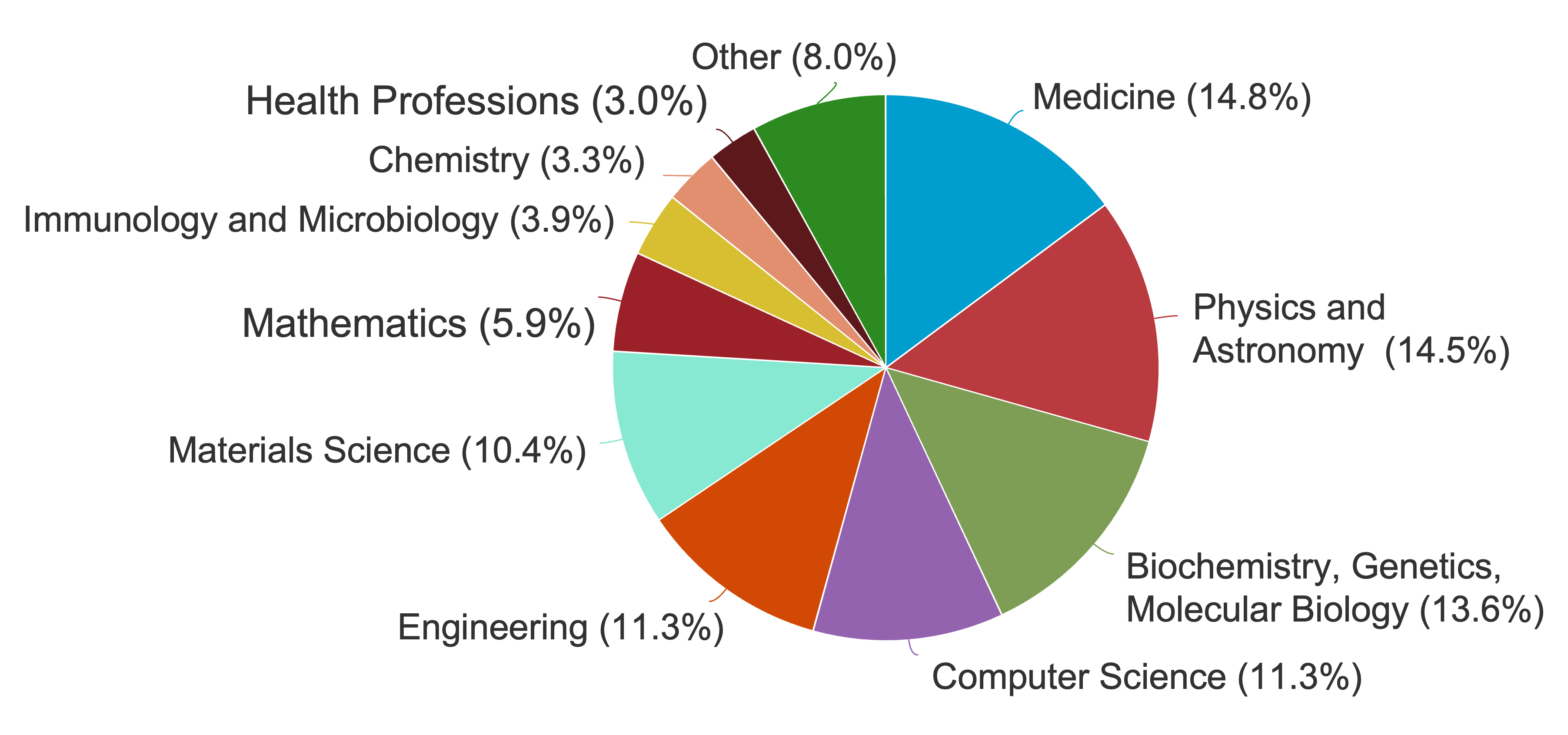
My areas of research according to Scopus:
Page updated on 2022-10-28 11:27:35 -0400